- Need assistance?
- 1-888-762-2568
- Contact Us
SeqPure PCR Purification Kit (5 ml)
- Applications: NGS purification
Need help with your order?
Find out more about placing an order here or contact us for technical support.Product Description
BioChain's SeqPure provides an efficient, convenient, and cost effective way to clean up DNA that is greater than 100bp in length.Product Specifications
| Features | Specifications |
|---|---|
| Size | 5 ml |
| Shipping | Ambient Temperature |
| Sex | Contact Us |
| Age At Sampling | Contact Us |
| Tissue | N.A. |
| Reagent Type | DNA Cleanup |
| Method | Manual, Automated |
| Applications | NGS purification |
Protocols
Flyers & Brochures
SDS
Certificate of Analysis
Applications:
• PCR
• Mutation detection and genotypying
• Sequencing (Sanger and Next Generation)
• Microarrays
• Restriction enzyme clean up
• Cloning
• Fragment analysis
Advantages and Features:
• Similar performance as leading competitors, at a budget-friendly price
• Purifies PCR products in under half an hour
• Removes salts, dNTPs, primers, and enzymes
• No centrifugation or vacuum filtration required
• No fancy buffers; just use ethanol and Tris-HCI/TE
• Easily automated
• Amenable to 96- and 384- well formats
• Scalable: Only use as many beads as you need
Excellent Recovery of DNA Fragments
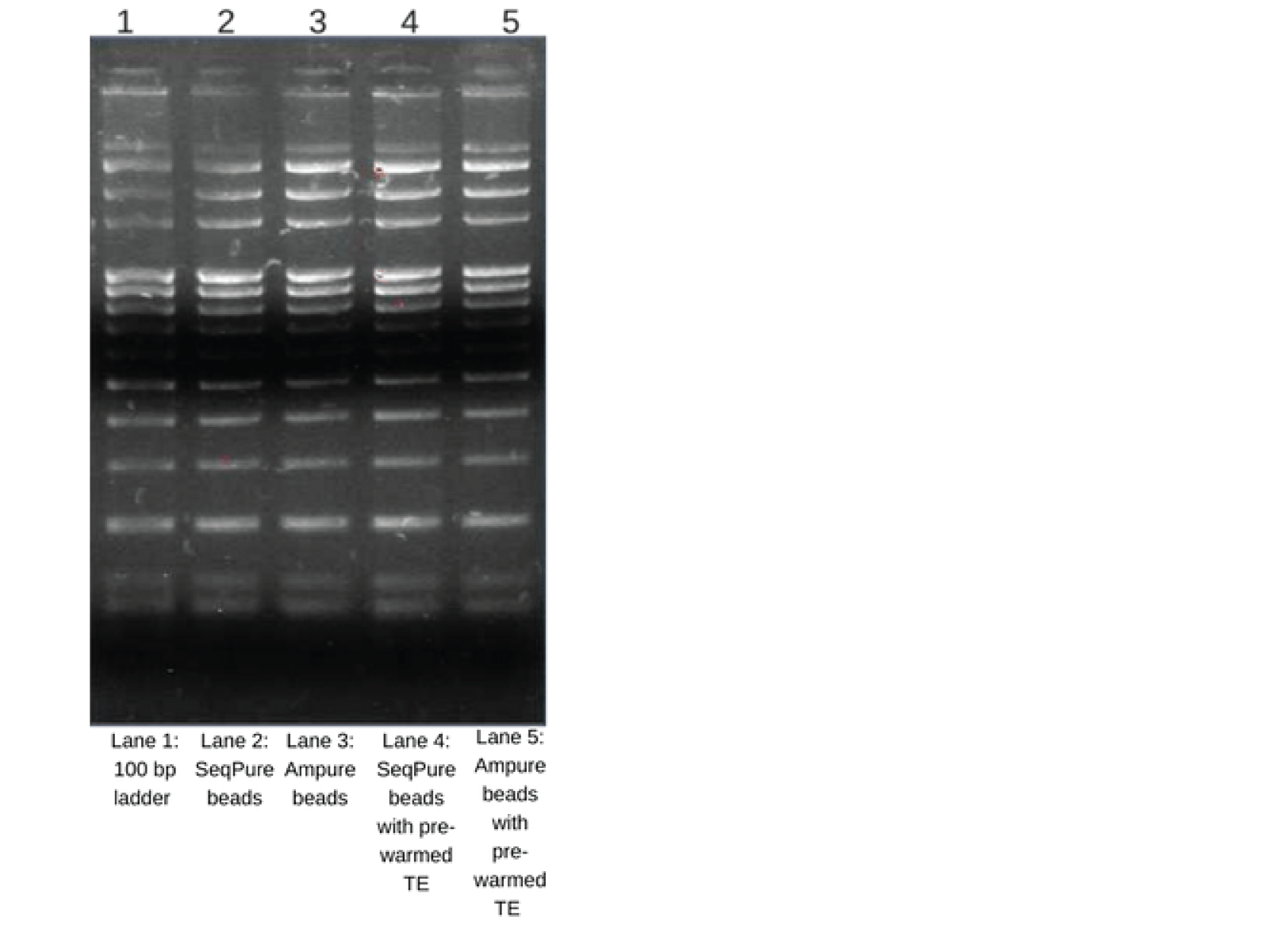
Equal amounts of 100 bp ladder containing salts and proteins were purified with SeqPure beads and Ampure beads. Different buffers, TE and pre-warmed TE (for increasing the yield) were used for elution. Eluted DNA (8 µl each) were loaded and run on a 1% agarose gel. The gel shows excellent recovery of all fragments from 3000-100bp.
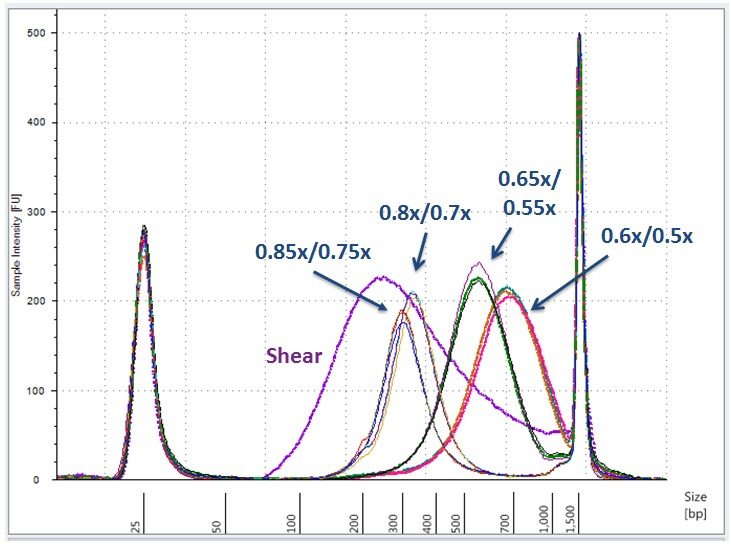
Efficient Size Selection
Sample Protocol
Excellent DNA Purification
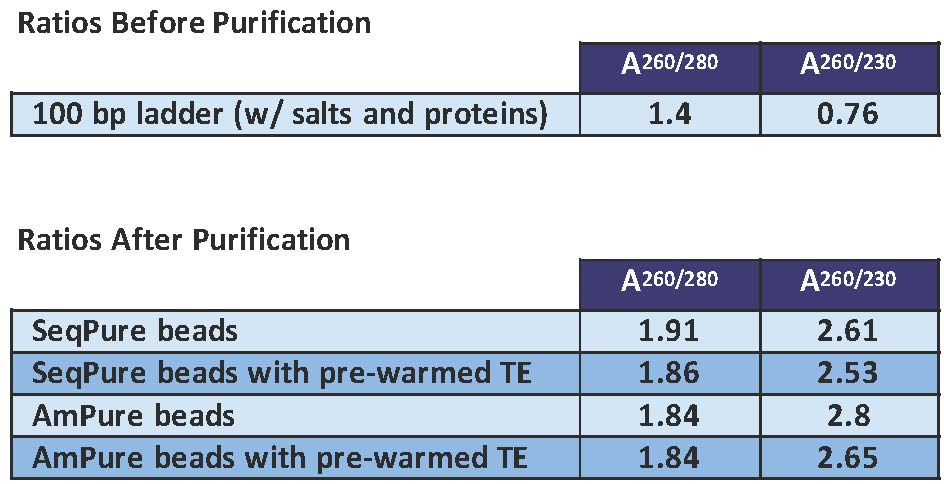
Comparison of SeqPure and AmPure in purifying 100 bp DNA ladder containing salts and protein. The purity was measured by spectrophotometry. Both the 260/280 and 260/230 absorbance ratios show that SeqPure is efficient at removing salts and proteins and is comparable to AmPure.
More Information About SeqPure PCR Purification Kit
For many molecular biology techniques, removing contaminants from DNA is critical for downstream applications. For example, to ensure a successful sequencing reaction, all excess contaminants such as primers, short-failed PCR products, unincorporated dNTPs, salts, and residual proteins, must be removed from the template. These agents can interfere with subsequent reactions and lead to unreadable sequences.
While removal of excess contaminants is key to producing the highest quality data, it can be time-consuming to do so. A PCR purification kit that makes DNA cleanup convenient can dramatically improve productivity in the lab. A kit that can be used manually for small sample numbers, while being easily automated on a variety of platforms, insures that the same procedure can be adopted for all sample throughputs – insuring consistent performance. Such a kit should also be efficient, and purify DNA fragments with minimal losses. Lastly, it should be cost-effective.
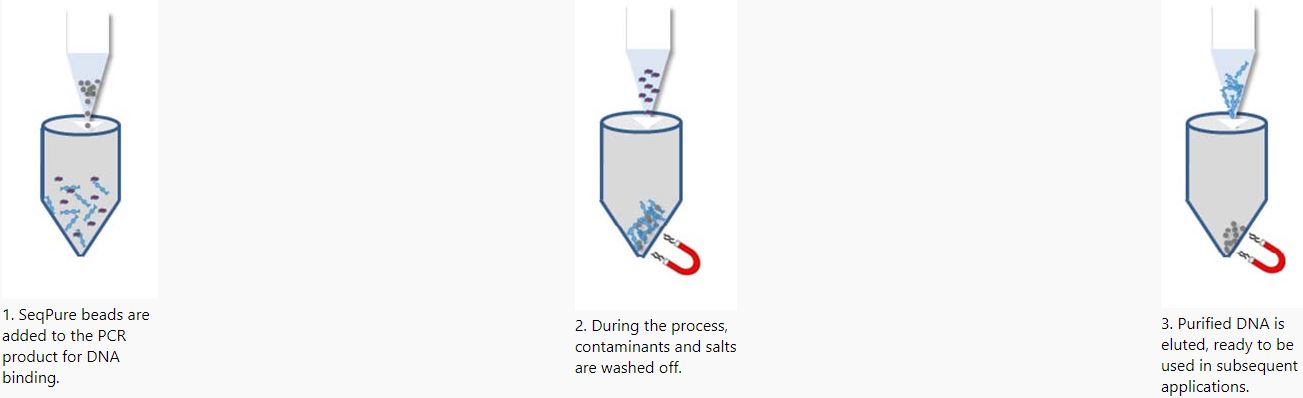
BioChain’s SeqPure PCR Purification kit is designed for rapid and efficient purification of PCR products. The system utilizes paramagnetic bead technology to selectively bind DNA fragments 100 bp and larger in size. Simple washing steps will remove salts and other buffer constituents, primers, primer dimers, nucleotides, and enzymes prior to elution of the purified PCR product. SeqPure avoids the use of spin-columns and other purification membranes in order to make the kit convenient to use manually or automate on the widest possible selection of laboratory automation (eg. KingFisher, Beckman, Hamilton, Tecan, Caliper, Perkin Elmer, Agilent and Eppendorf). 96- and 384- well formats can easily be accommodates with this technology. Purification with paramagnetic particles, rather than spin columns reduces hydrodynamic shearing – improving the integrity of larger DNA fragments. The scalable nature of paramagnetic bead technology also makes the kit more flexible. If you are purifying small amounts of DNA, you can use fewer beads, and extract more samples per purification kit.