
Reverse Transcription: Its Discovery and Existence
Reverse transcription is the process of generating cDNA (complementary DNA) from mRNA and the enzyme catalyzing this process is called reverse transcriptase (RT).
It was during the 1970s that two scientists named Temin and Baltimore discovered independently the RT (RNA- and DNA-dependent DNA polymerase) (Baltimore, D., 1970; Temin and Mizutani, 1970) enzyme in viruses, for which they won the Nobel Prize. They wanted to study the virus-cancer relationship and found that there was an enzyme that produced a DNA intermediate.
Reverse Transcription: Its Function
Reverse transcriptase consists of 2 domains:
- DNA polymerase – extends nucleotide in 5′ → 3′ direction
- RNase H - an endonuclease which hydrolyses RNA when present in an RNA-DNA duplex (Sevilya et al., 2003).
Do only retroviruses, which composed of RNA and not DNA, have the RT enzyme? The simple answer is no. RT enzyme is not restricted to retroviruses (Hull R., 1999)
Retroelements, which are key genetic elements in retroviruses, carry information from one location to another within the genome through reverse transcription of an RNA intermediate and are classified into 3 groups:
- viral (retrovirus and pararetrovirus)
- bacterial (retron), and
- eukaryotic non-viral retroelements (retrotransposon and retroposon, all of which contains the RT enzyme.
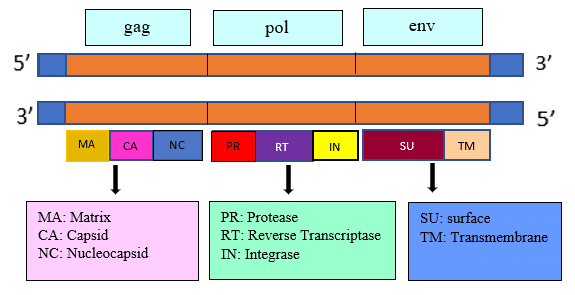
Let us look specifically into the retroviral genome, which has RNA as its genetic material and concentrate on three important genes encoding viral proteins. Above Right: Location of gag, pol, and env genes in retroviral genome. Gag region constitutes the group antigens and make up the major core structure. Pol region contains protease, reverse transcriptase, and integrase enzymes. Env region composes of transmembrane and surface proteins surrounding the core.
Popular RT’s
Among all RT’s, AMV (Avian Myeloblastosis Virus) and MMLV (Moloney Murine Leukemia Virus) are most preferred in the laboratories and have the following properties.
TABLE I Right: Comparison between AMV and MMLV RT. Processivity is the number of nucleotides added to generate cDNA, thermostability describes the ability of RT to tolerate high temperatures, and fidelity is used to describe the accuracy of the polymerase to correctly synthesize new DNA strand

Which RT Should be Used?
Each of the two RT’s has its specific characteristics, with advantages and disadvantages. Currently, most researchers prefer the mutant form of MMLV RT or a combination of both of the above.
Applications of RT:
- Reverse Transcription PCR (RT-PCR), quantitative (qPCR)
- microarray study
- probe generation
- cDNA library preparation (BioChain has a large collection of cDNA made from various species)
- RNA sequencing experiments to study alternative splicing, gene expression profiling, SNP identification, assist in the development of antibodies, disease association study, and to annotate new transcripts.
Limitations of RT:
- It does not have 3′-5′ exonuclease activity, which is required for proofreading activity. Hence, mutation/error rates are high during cDNA synthesis and lead to low fidelity.
- Due to its sensitivity towards alcohol, phenol, detergents or salt used during the isolation of RNA, it leads to non-uniformity towards the PCR reaction used (Freeman et al., 1999)
- Changes in the bases such as 2′‐O‐methylated nucleotides can restrict the RT enzyme (Kellner et al., 2010)
- Owing to its RNase H activity, truncated cDNA is produced as the RNA molecules get hydrolysed in a DNA-RNA hybrid form (Boone et al., 2018)
- On encountering secondary structure, the enzyme halts, unable to go through full-length RNA.
- At elevated temperatures, RT loses its thermostability, posing a hindrance in cDNA synthesis (Baranauskas et al., 2012)
BioChain: Ease and Convenience
BioChain offers most popularly used enzymes including the wild-type (MMLV RT) and the recombinant MMLV RT (UltraScript MMLV RT) purified from E. coli. The recombinant form exhibits higher thermostability and lower RNase H activity, allowing higher cDNA yield and full-length cDNA with small amount of total RNA (10 pg to 5 µg). It works well within 37o C to 60o C range and the higher temperatures make it possible to deal with templates having secondary structure. The high-quality RT comes with a 5X reaction buffer providing the advantage to choose one’s protocol suitable for several downstream analyses.
BioChain combines leading technology along with good laboratory practices (GLP) to ensure releasing the best product in the market. It is a one-stop place for scientists and customers offering high-quality products with competitive pricing.

References
- Baltimore, D. (1970). Viral RNA-dependent DNA Polymerase: RNA-dependent DNA Polymerase in Virions of RNA Tumour Viruses. Nature, 226(5252), 1209-1211. doi: 10.1038/2261209a0.
- Temin, H., and Mizutani, S. (1970). Viral RNA-dependent DNA Polymerase: RNA-dependent DNA Polymerase in Virions of Rous Sarcoma Virus. Nature, 226(5252), 1211-1213. doi: 10.1038/2261211a0
- Sevilya, Z., Loya, S., Adir, N., and Hizi, A. (2003). The ribonuclease H activity of the reverse transcriptases of human immunodeficiency viruses type 1 and type 2 is modulated by residue 294 of the small subunit. Nucleic Acids Research, 31(5), 1481-1487. doi: 10.1093/nar/gkg235
- Hull, R. (1999). Classification of reverse transcribing elements: a discussion document. Archives Of Virology, 144(1), 209-214. doi: 10.1007/s007050050498Freeman, W., Walker, S., and Vrana, K. (1999). Quantitative RT-PCR: Pitfalls and Potential. Biotechniques, 26(1), 112-125. doi: 10.2144/99261rv01
- Kellner, S., Burhenne, J., and Helm, M. (2010). Detection of RNA modifications. RNA Biology, 7(2), 237-247. doi: 10.4161/rna.7.2.11468
- Boone, M., De Koker, A., and Callewaert, N. (2018). Capturing the ‘ome’: the expanding molecular toolbox for RNA and DNA library construction. Nucleic Acids Research, 46(6), 2701-2721. doi: 10.1093/nar/gky167
- Baranauskas, A., Paliksa, S., Alzbutas, G., Vaitkevicius, M., Lubiene, J., and Letukiene, V. et al. (2012). Generation and characterization of new highly thermostable and processive M-MuLV reverse transcriptase variants. Protein Engineering Design And Selection, 25(10), 657-668. doi: 10.1093/protein/gzs034
Abbreviations
RT, Reverse Transcriptase; AMV, Avian Myeloblastosis; MMLV, Moloney Murine Leukemia Virus
Author
Sujata Dhar

