
Liquid Biopsies
Liquid biopsies offer a minimally invasive method for effective cancer detection and prognosis in the form of circulating biomarkers (Cree 2015). Though the exclusivity and conclusivity of liquid biopsies as the most efficacious forms of cancer prognosis and treatment remains doubtful, there is no second thought to admitting its importance in the identification of therapeutically relevant genetic mutations leading to the most dangerous and fatal tumors (Jung and Kirchner 2018).
Circulating Nucleic Acids in Liquid Biopsy Samples
Liquid biopsies involve examination of circulating tumor cells (CTCs), cell-free DNA (cfDNA) and circulating-tumor DNA (ctDNA) as a measure to gain information on tumor molecular genetics, monitor the extent of tumor growth (especially in the case of suspected metastases), check for chromosomal aberrations, mutations and track mechanisms of drug resistance in solid tumors(Economopoulou et al. 2017).
Early diagnosis and better treatment outcomes have been the major areas of focus for cancer researchers for decades, and this quest has found a formidable answer in the form of ctDNA and cfDNA biomarkers, which when analysed via high throughput sequencing methods, can give considerable information on the tumors at a genomic level – something that is extremely unique for every individual (Cree et al. 2017).
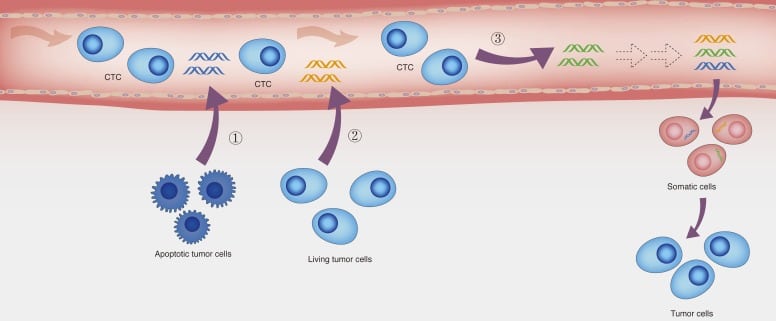
Why cfDNA / ctDNA?
It is a known fact that all individuals carry circulating DNA in their blood in the form of low levels of cfDNA. The circulating cfDNA is believed to be released by necrotic cells due to cell-death. Other studies have also indicated that cfDNA is derived from active cellular secretions such as exosomes, apoptotic blebs, shedding vesicles, and microparticles (Wang et al. 2017). In case of any disorder or pathogenic condition (e.g. myocardial infarction, infectious diseases and inflammatory conditions, different kinds of cancers and even pregnancy) this number goes up (Cree 2015). In the case of patients with cancers or growing tumors, the cell-free DNA circulating in bloodstream is referred to as ctDNA (Cree et al. 2017).
Rising Popularity of ctDNA Testing
ctDNA testing and analysis has found tremendous potential in testing for breast cancer, different types of squamous cell carcinomas, gastrointestinal cancers, prenatal genetic testing and testing for immunological aberrations in a donor’s system post organ-transplantation (Volik et al. 2016). Checking for tumor-specific genes with the help of specific primers have earlier yielded significantly positive results in patients with pancreatic adenocarcinoma and acute myelogenous leukemia (AML) and with the advent of molecular genetics, one can only hope for further improvements in genetic tumor testing.
Existing Methods and Challenges
ctDNA can be measured and analysed by a number of methods, the most common methods being BEAMing, PCR clamping methods, and deep sequencing using NGS (Cree et al. 2017). Sensitivity and cost are two parameters which have affected researchers and scientists in their hunt for the ‘perfect’ method to diagnose cancers early. Some of the factors contributing to challenges in sensitive and robust ctDNA testing include low amount, high degradation, and high mixture of normal DNA in cfDNA (Volik et al. 2016).
What BioChain has to offer?
Keeping the current scenarios in mind, Biochain offers a range of products which help in optimal utilization of the available sample for extraction of nucleic acids. These products make analysis of circulating nucleic acids easier, faster and more specific in nature.
BioChain’s cfPure® & cfPure® MAX Cell Free DNA Extraction Kits are designed for rapid and efficient purification of circulating cell free DNA (cfDNA).
It offers its users rapid and easy processing of multiple samples in an hour or less making it ideal for biomarker screening. BioChain has developed its own silica-coated magnetic bead technology, which has been designed to efficiently recover low molecular weight cell free DNA during isolation. Not only this, the kit is also scalable and one can decide on the exact amount of sample necessary to execute an effective diagnosis.
Once cfDNA is extracted, products of downstream applications such as PCR and NGS library prep can be cleaned up using BioChain’s SeqPure™ PCR Purification system. SeqPure™ is a post PCR and Next Gen library prep clean up system also based on paramagnetic beads technology, designed for rapid and efficient purification of PCR amplicons.
References
- Cree, Ian A. 2015. “Liquid Biopsy for Cancer Patients: Principles and Practice.” Pathogenesis 2 (1): 1–4. https://doi.org/10.1016/j.pathog.2015.05.001.
- Cree, Ian A., Lesley Uttley, Helen Buckley Woods, Hugh Kikuchi, Anne Reiman, Susan Harnan, Becky L. Whiteman, et al. 2017. “The Evidence Base for Circulating Tumour DNA Blood-Based Biomarkers for the Early Detection of Cancer: A Systematic Mapping Review.” BMC Cancer 17 (October). https://doi.org/10.1186/s12885-017-3693-7.
- Economopoulou, P., I. Kotsantis, E. Kyrodimos, E. S. Lianidou, and A. Psyrri. 2017. “Liquid Biopsy: An Emerging Prognostic and Predictive Tool in Head and Neck Squamous Cell Carcinoma (HNSCC). Focus on Circulating Tumor Cells (CTCs).” Oral Oncology 74 (November): 83–89. https://doi.org/10.1016/j.oraloncology.2017.09.012.
- Jung, Andreas, and Thomas Kirchner. 2018. “Liquid Biopsy in Tumor Genetic Diagnosis.” Deutsches Ärzteblatt International 115 (10): 169–74. https://doi.org/10.3238/arztebl.2018.0169.
- Volik, Stanislav, Miguel Alcaide, Ryan D. Morin, and Colin Collins. 2016. “Cell-Free DNA (CfDNA): Clinical Significance and Utility in Cancer Shaped By Emerging Technologies.” Molecular Cancer Research 14 (10): 898–908. https://doi.org/10.1158/1541-7786.MCR-16-0044.
- Wang, Wei, Peng Kong, Ge Ma, Li Li, Jin Zhu, Tiansong Xia, Hui Xie, Wenbin Zhou, and Shui Wang. 2017. “Characterization of the Release and Biological Significance of Cell-Free DNA from Breast Cancer Cell Lines.” Oncotarget 8 (26): 43180–91. https://doi.org/10.18632/oncotarget.17858.
Author
BioChain Institute Inc.

