Introduction to Biopsy
Q: What is tissue and liquid biopsy?
A: Biopsy is a surgical procedure to determine the presence of cancerous cells in our body. Tissue biopsy involves taking out a piece of tissue, examining it under a microscope and then making decision if the patient has a disease. A liquid biopsy involves taking a sample of fluids, mostly blood, from the patient and test for any sign of abnormality.
Q: What is FFPE and why is it needed?
A: It is a technique of fixing the tissue in formalin, followed by embedding them in paraffin blocks. Hence, the name is formalin-fixed paraffin-embedded (FFPE). It is mainly used as an archive and stored in biobanks for preservation, later to be used in clinical diagnosis, microscopic analysis, and personalized medicine.

Figure 1: Preparation of FFPE tissue blocks
Pros and Cons of using FFPE
Pros:
- Convenient storage system- Room temperature
- Clinically validated and remains a gold standard still today
- Used for retrospective study
- Preserves tissue from degrading
- Keeps cells and its contents intact
- Availability of valuable clinical information
- Helps to analyze various histological cancer properties
Cons:
- Invasive procedure
- Gives an overview of only the tissue removed
- Hard to look for disease progression
- Lack of tumour heterogeneity
- Less flexibility of repetition
- Creates bias as only a selected population of the tumor is represented
- Artifactual mutations
Standardization for FFPE Sample Storage
The US Office of Biorepositories and Biospecimen Research of National Cancer Institute (NCI) have set guidelines for nucleic acid extraction from FFPE samples (Greytak et al.) and European Committee for Standardization Technical Specifications (CEN/TS) have recommendations on the pre-analytical phase of FFPE (Kofanova et al.)
Below are certain criteria that needs to be inspected in order to extract DNA:
- Timing for tissue ischemia and fixation
- Temperature and storage conditions
- Methods used to extract DNA- types of kits and assays
In spite of the guidelines, there still exists variability in the FFPE tissue processing, based on how, when, and what techniques are being used to prepare the blocks. Quality control steps and data interpretation can also vary a lot depending on the standards set by an institution or a company.
Application of FFPE in NGS
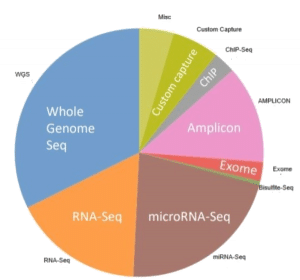
The quality of FFPE tissues is of utmost important, which can be a make-or-break in analytical steps. That is why, BioChain uses stringent methods on its FFPE tissues. ( Figure 2: Use of FFPE in different NGS applications, illustrated by a pie-chart.)
- Predominantly used in cancer biology as biomarkers
- Creating patient stratification cohort study (Pennock et al.)
- Study host immune response in patients (Conroy et al.)
- Gene fusion identification, gene expression, and variant detection (Norton et al.)
- Detection of actionable mutation (mutation responsive of a treatment)
BioChain’s prime-quality CancerSeq™ FFPE Tissues
These tissues have been pre-screened for the following:
- Single nucleotide polymorphisms (SNPs)
- Insertions/deletions (indels)
- Copy number variations (CNVs)
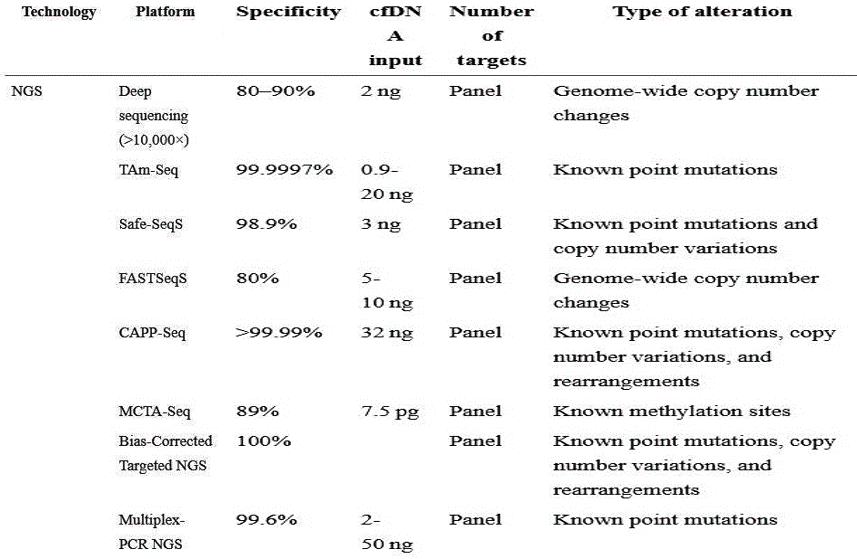
- RNA fusions
DNA extracted from FFPE tissues was screened for mutations with Next Generation Sequencing (NGS) using Illumina’s TruSeq Amplicon - Cancer Panel (TSACP), ArcherDX’s VariantPlex Solid Tumor Panel which include 48 or 67 cancer-related genes or Thermo Fisher’s Ion AmpliSeq panel. (Figure 3: BioChain’s HE stained tumor matched pair tissue sections; A: Paraffin embedded breast cancer tissue B: Paraffin embedded breast cancer adjacent normal tissue)

Features:
- Includes variant calling by application of stringent methods
- Deep sequencing with reduced false positives
- Has 67 cancer gene panels
- Detailed clinical history available of the tissue donors
- Data provided on NGS parameters- quality score, depth, variant types and so on.
Applications:
- Identification of novel biomarker
- Validation for mutation studies
- Suited for Immunohistochemistry (IHC) and In- situ hybridization (ISH) assays
- Cellular localization of tissue-specific mRNA and protein expression
- Companion diagnostics assay development
- Controls or verification for genotyping
Replacement to Tissue Biopsy?
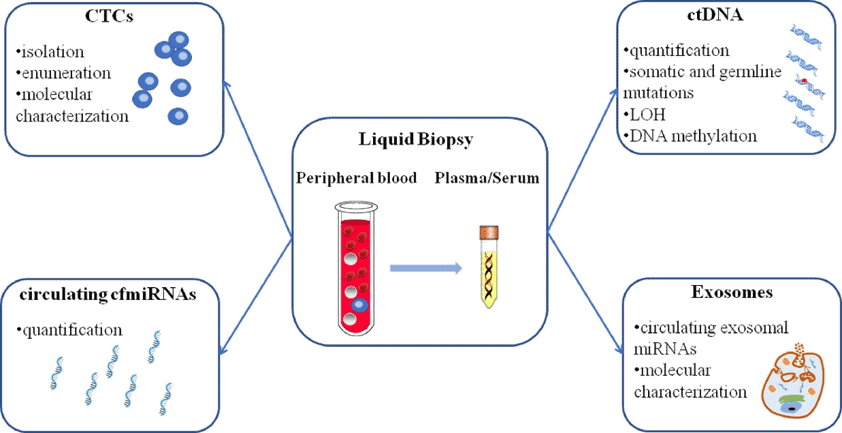
Liquid biopsy could be a potential replacement to tissue biopsy for many researchers. ( Figure 4: Different approaches of liquid biopsy such as using Circulating tumor cells (CTCs), circulating tumor DNA (ctDNA), circulating cell-free microRNAs (cfmiRNAs), and exosomes.) Among them, circulating cell-free DNA (cfDNA) has been receiving special attention in research. Let us have a look why.
What is cell-free DNA (cfDNA)?
circulating tumor DNA (ctDNA) - originating from the tumor cells, consists of a small fraction of the cfDNA
It is found in the body fluids, such as, blood, urine, saliva, cerebrospinal fluid, and breast milk as DNA fragments. They are present in the circulation, representing the remnants of tumor, and hence can act as cancer biomarkers. Studies have indicated high level of cfDNA in the plasma of cancer patients compared to the healthy control (Temilola et al.). Q: Is there any standardization developed for circulating nucleic acids? A: Yes, there are being initiatives taken. CANCER-ID is a public-private project aimed at standardization of protocols starting with sample collection to storage, characterization, and validation of assays for circulating nucleic acids. The Blood Profiling Atlas in Cancer (BloodPAC) Consortium , is also set out to promote the research in liquid biopsy.
Pros:
- Time to process FFPE samples is more
- Blood draw is easier as compared to multiple biopsies
- Sample size for tissue is limited
- Preservation is easier
- Helps to monitor serial progression of disease
- Provides real-time picture of the disease- the half-life of cfDNA in the circulation is 16 mins-2.5 hr (Chen and Zhao)
- Representative of tumor heterogeneity
- Helps to assess the risk of relapse in the patients
Cons:
- Sensitivity is low compared to tissue biopsy
- Detects mutation from hematopoietic clones instead of the actual tumor
- Proper clinical guidelines are not yet established
- Possibility of false positives- detection of cancer, when in reality it is not present (“Biopsy: Using DNA in Blood to Detect, Track, and Treat Cancer”)
- May be misleading in making decisions- the release of ctDNA is representative of whole or part of the tumor
- Distinguishing between carrier and passenger mutation of ctDNA is difficult
Liquid Biopsy Study
The Circulating Cell-Free Genome Atlas (CCGA) was the largest study undertaken, aimed at developing a liquid biopsy to screen patients with lung cancer in the US with varying clinical histories. The cfDNA was isolated and three sequencing methods were applied. All of these methods detected lung cancers and observed that the sensitivity increased with increased cancer stage
Application of ctDNA in NGS
- Deep sequencing of multiple genes
- Used in targeted gene panels to detect mutations
- Screen for fetal chromosomal abnormalities – a test called non-invasive prenatal testing (NIPT)
- Understanding the relationship between microbe and human health conditions (Chiu and Yu)
- Monitoring lung transplant patients for any risks involved
- Microsatellite Instability detection (Lozac’hmeur et al.)
- Gather insights about cancer and its progression
- Examine the tumor response to drug therapy in patients (Frenel et al.)
- Use as a Companion Diagnostics such as Cobas EGFR Mutation Test v2
BioChain’s Cell Free DNA Extraction Kit
cfPure® Cell Free DNA Extraction Kit is a silica-coated magnetic bead-based extraction kit with high recovery rate. It has simple, quick, and high-yielding protocol to ensure the sample is of top-notch quality. The added advantage is that compared to other column-based kits available, cfPure® can match the gold standard in terms of yield and purity but more scalable, economic, and automation friendly. It is high specificity and can be used in sensitive applications, such as Next Generation Sequencing, to all the current spin column based cfDNA kits available in the market.

Figure 5. cfDNA was purified from 2 mL of plasma from different donors by either the cfPure® Cell Free DNA Extraction Kit or by a competitor’s DNA extraction kit. DNA recovery was assessed either by a fluorescent method
To read more on different metrics and parameters, where BioChain’s cfPure® kit outperformed compared to other kits, click on the following link. Click here for more info.
Future of Tissue Biopsy (FFPE) and Liquid Biopsy (cfDNA) in NGS
While storage of FFPE tissues are convenient, providing an ample source of information for the researchers, it has its limitations when interpreting NGS data.
Here are few:
- The storage time can impact the quality of nucleic acid.
- The use of formalin produces artificial C: G>T: A mutation. Due to its occurrence in normal tissues, this results in false positive results. Treating the tissue with Uracil DNA glycosylase (UDG) can help eliminate this bias (Kim et al.)
- Non-uniformity is observed in the sequencing data. The way to address this is to increase the coverage for sequencing
- Quality Control (QC) metric for RNA-Seq data such as RNA integrity number (RIN) does not hold accurate information. DV100- percentage of RNA fragments having more than 100 nucleotides is found to be more informative
- Heavily degraded RNA can have 3’ bias. It has been found that rRNA depletion and random priming during reverse transcription yields good sequencing libraries (Zhao et al.)
- Lack of matched fresh frozen tumor sample from the same patient for comparison
Nonetheless, scientists have successfully conducted Whole Genome (WG) and Whole Exome (WE) studies using FFPE samples.
It is so much convenient to isolate cfDNA without having to think of cost or inducing pain to the patient. However, ctDNA is only 0.01% of cfDNA, which makes the task of detecting the early stage cancer challenging (Elazezy and Joosse). Lack of standardization for handling ctDNA will make the diagnosis even tougher. In addition to tumor cells, normal and hematopoietic cells also release cfDNA, thus contributing to false-positive tests, when analysing ctDNA in cancer (Chen and Zhao). These are few drawbacks in ctDNA sequencing.
However, it can be a powerful tool to look for tumor in order to avoid the risk of tumor rupture and worsening the condition. To know that the concentration of ctDNA differs from high risk patient to low risk patient and healthy versus diseased can be enlightening to be used as a biomarker for the future (Van Paemel et al.)
Thus, tissue biopsy is still going to be the gold standard, while liquid biopsy can be used as an added resource for supporting the main findings.
References
- “Biopsy: Using DNA in Blood to Detect, Track, and Treat Cancer.” Biopsy: Using DNA in Blood to Detect, Track, and Treat Cancer, 8 Nov. 2017, www.cancer.gov/news-events/cancer-currents-blog/2017/liquid-biopsy-detects-treats-cancer. Accessed 21 Mar. 2020.
- Chen, Ming, and Hongyu Zhao. “Next-Generation Sequencing in Liquid Biopsy: Cancer Screening and Early Detection.” Human Genomics, vol. 13, no. 1, 1 Aug. 2019, 10.1186/s40246-019-0220-8. Accessed 17 Oct. 2019.
- Chiu, Kuo-Ping, and Alice L. Yu. “Application of Cell-Free DNA Sequencing in Characterization of Bloodborne Microbes and the Study of Microbe-Disease Interactions.” PeerJ, vol. 7, 6 Aug. 2019, p. e7426, 10.7717/peerj.7426. Accessed 22 Mar. 2020.
- Conroy, Jeffrey M., et al. “Analytical Validation of a Next-Generation Sequencing Assay to Monitor Immune Responses in Solid Tumors.” The Journal of Molecular Diagnostics, vol. 20, no. 1, Jan. 2018, pp. 95–109, 10.1016/j.jmoldx.2017.10.001. Accessed 21 Mar. 2020.
- Elazezy, Maha, and Simon A. Joosse. “Techniques of Using Circulating Tumor DNA as a Liquid Biopsy Component in Cancer Management.” Computational and Structural Biotechnology Journal, vol. 16, 2018, pp. 370–378, www.ncbi.nlm.nih.gov/pmc/articles/PMC6197739/, 10.1016/j.csbj.2018.10.002. Accessed 6 Nov. 2019.
- Frenel, Jean Sebastien, et al. “Serial Next Generation Sequencing of Circulating Cell Free DNA Evaluating Tumour Clone Response To Molecularly Targeted Drug Administration.” Clinical Cancer Research : An Official Journal of the American Association for Cancer Research, vol. 21, no. 20, 15 Oct. 2015, pp. 4586–4596, www.ncbi.nlm.nih.gov/pmc/articles/PMC4580992/, 10.1158/1078-0432.CCR-15-0584. Accessed 22 Mar. 2020.
- Greytak, Sarah R., et al. “National Cancer Institute Biospecimen Evidence-Based Practices: Harmonizing Procedures for Nucleic Acid Extraction from Formalin-Fixed, Paraffin-Embedded Tissue.” Biopreservation and Biobanking, vol. 16, no. 4, Aug. 2018, pp. 247–250, 10.1089/bio.2018.0046. Accessed 21 Mar. 2020.
- Kim, Seokhwi, et al. “Deamination Effects in Formalin-Fixed, Paraffin-Embedded Tissue Samples in the Era of Precision Medicine.” The Journal of Molecular Diagnostics, vol. 19, no. 1, Jan. 2017, pp. 137–146, 10.1016/j.jmoldx.2016.09.006. Accessed 22 Mar. 2020.
- Kofanova, Olga, et al. “Standardization of the Preanalytical Phase of DNA Extraction from Fixed Tissue for Next-Generation Sequencing Analyses.” New Biotechnology, vol. 54, 25 Jan. 2020, pp. 52–61, www.sciencedirect.com/science/article/pii/S1871678419300469#bib0050, 10.1016/j.nbt.2019.07.005. Accessed 21 Mar. 2020.
- Lozac’hmeur, Ariane, et al. “Microsatellite Instability Detection with Cell-Free DNA next-Generation Sequencing.” The Society for Immunotherapy of Cancer, 6 Nov. 2019.
- Nagahashi, Masayuki, et al. “Formalin-Fixed Paraffin-Embedded Sample Conditions for Deep next Generation Sequencing.” The Journal of Surgical Research, vol. 220, 1 Dec. 2017, pp. 125–132, www.ncbi.nlm.nih.gov/pmc/articles/PMC5726294/, 10.1016/j.jss.2017.06.077.
- Norton, Nadine, et al. “Gene Expression, Single Nucleotide Variant and Fusion Transcript Discovery in Archival Material from Breast Tumors.” PLoS ONE, vol. 8, no. 11, 22 Nov. 2013, p. e81925, 10.1371/journal.pone.0081925. Accessed 21 Mar. 2020.
- Pennock, Nathan D., et al. “RNA-Seq from Archival FFPE Breast Cancer Samples: Molecular Pathway Fidelity and Novel Discovery.” BMC Medical Genomics, vol. 12, no. 1, Dec. 2019, 10.1186/s12920-019-0643-z. Accessed 21 Mar. 2020.
- Temilola, Dada Oluwaseyi, et al. “The Prospect and Challenges to the Flow of Liquid Biopsy in Africa.” Cells, vol. 8, no. 8, 1 Aug. 2019, p. 862, www.mdpi.com/2073-4409/8/8/862/htm, 10.3390/cells8080862. Accessed 22 Mar. 2020.
- Van Paemel, Ruben, et al. “The Pitfalls and Promise of Liquid Biopsies for Diagnosing and Treating Solid Tumors in Children: A Review.” European Journal of Pediatrics, vol. 179, no. 2, 3 Jan. 2020, pp. 191–202, 10.1007/s00431-019-03545-y. Accessed 22 Mar. 2020.
- Zhao, Yongmei, et al. “Robustness of RNA Sequencing on Older Formalin-Fixed Paraffin-Embedded Tissue from High-Grade Ovarian Serous Adenocarcinomas.” PLOS ONE, vol. 14, no. 5, 6 May 2019, p. e0216050, 10.1371/journal.pone.0216050. Accessed 22 Mar. 2020.
Abbreviations
Formalin-fixed paraffin-embedded, FFPE; National Cancer Institute, NCI; European Committee for Standardization Technical Specifications, CEN/TS; Single nucleotide polymorphisms, SNPs; Insertions/deletions, indels; Copy number variations, CNVs; Next Generation Sequencing, NGS; Immunohistochemistry, IHC; In- situ hybridization, ISH; Circulating tumor cells, CTCs; circulating tumor DNA, ctDNA; circulating cell-free DNA, cfDNA; The Circulating Cell-Free Genome Atlas, CCGA; noninvasive prenatal testing, NIPT; Uracil DNA glycosylase, UDG; Quality Control, QC; RNA integrity number, RIN; Whole Genome, WG; Whole Exome,WE
Author
BioChain Institute Inc.

