The Power of Multiomics
Multiomics and spatial multiomics analyses have become critical methods in studying tumor samples. Considering the spatial proximity of cells within a tissue sample alongside differential expression analysis offers a more comprehensive picture of disease development, key driver mutations, and possible therapeutic targets. (Multiomics Analysis Summary)
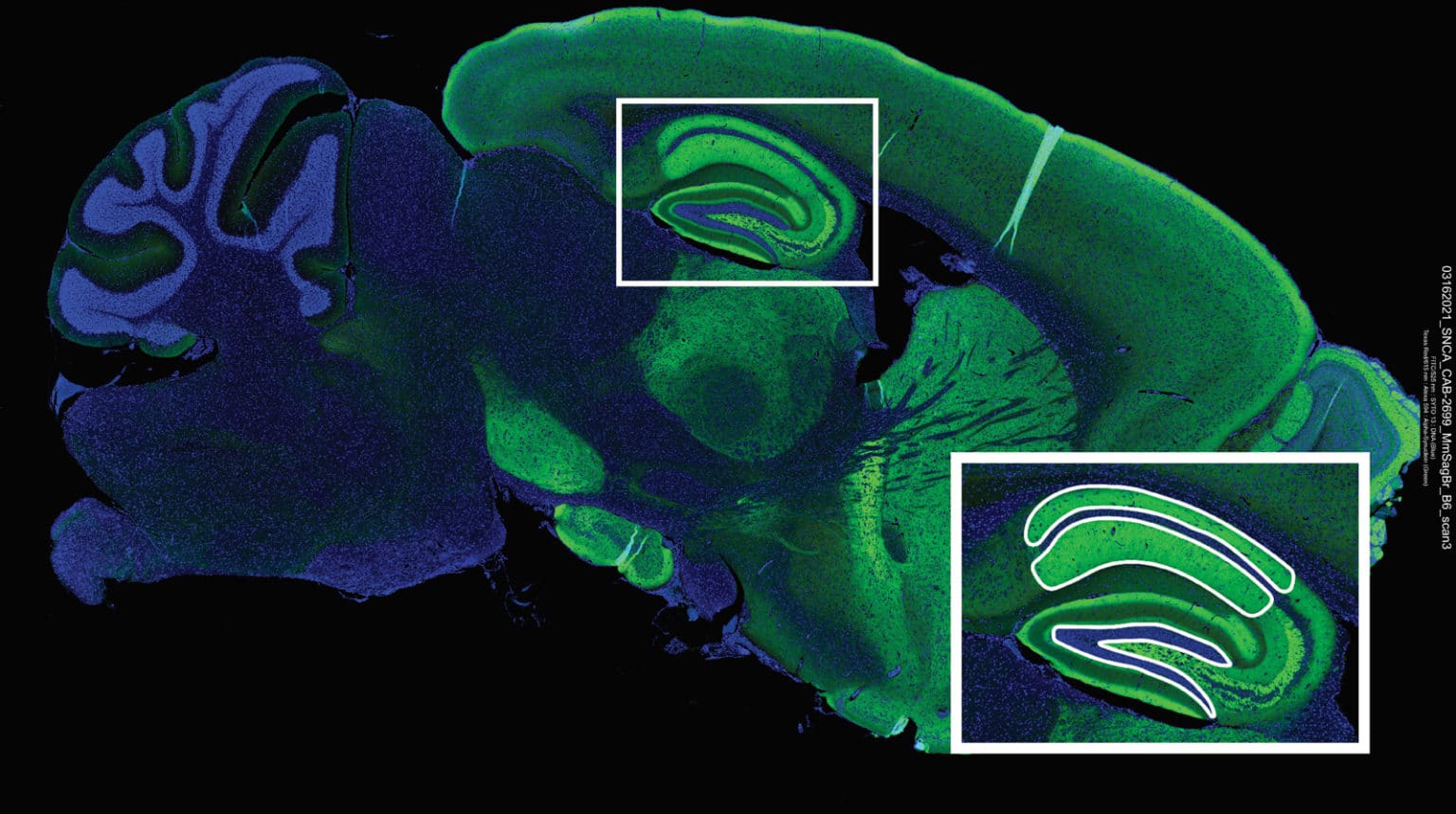
A 2021 publication by Rico et al. demonstrates the power of multiomics profiling.1 The study aim was to identify potential gene drivers and unique signaling pathways of DGASTs. DGASTs have been reported to occur in submucosal Brunner’s glands (BGs) of the duodenum.2 BGs however do not express GI hormones, and so DGASTs located in these glands must have a different tumor driver than found in other GI tumors.1
FFPE tissues (both tumor and normal) were analyzed by: (1) RNA sequencing for differential gene expression; (2) whole exome sequencing for SNPs and indels; (3) immunohistochemistry (IHC) for expression and cellular localization of proteins of interest; (4) Digital Spatial Profiling (DSP) (NanoString) to view expression patterns in spatial context, i.e. tumor cells and cells proximal that may provide clues to tumorigenesis; (5) qPCR for confirmation of differential gene expression.
This spatial multiomics study led to a hypothesis for the mechanism driving the development of DGASTs from normal tissue, as well as a potential therapeutic not previously considered for this disease subtype:
The Importance of Tissue Controls
As with any comparative study, utilizing proper control groups is critical. Since DGASTs are rare tumors, samples in this study were limited, and adjacent normal tissue was not available. To resolve this issue, normal tissue was obtained from various sources, including normal pancreas and duodenum from BioChain (Newark, California).
Multiomics Profiling
RNA sequencing followed by differential expression analysis was used to identify genes with high expression in DGASTs vs. other GI tumors. Once target genes were identified, IHC, qPCR and DSP were performed to explore differential expression in BG vs. DGAST tumors.
One particular gene of interest (NKX6.3) was found by IHC and qPCR to be expressed in DGAST cells but not in normal tissue – suggesting it plays a role in tumorigenesis or precursor events in the tissue prior to tumor formation. (For a one-step quantitative detection method for gene expression, consider BioChain’s QCell-Eva One-Step qRT-PCR SuperMix Kit.)
FFPE samples were analyzed with DSP to quantify a 40 target panel of neural-related antibodies and tumor morphology. DSP enables “visualization” of spatial context in samples that is otherwise missed by bulk RNA sequencing. In this study, DSP helped to identify pathways likely involved in the reprogramming of normal BG cells into DGAST tumors. This was possible by comparing expression patterns in stromal cells surrounding the tumors with cells of similar tissues to identify unique patterns.
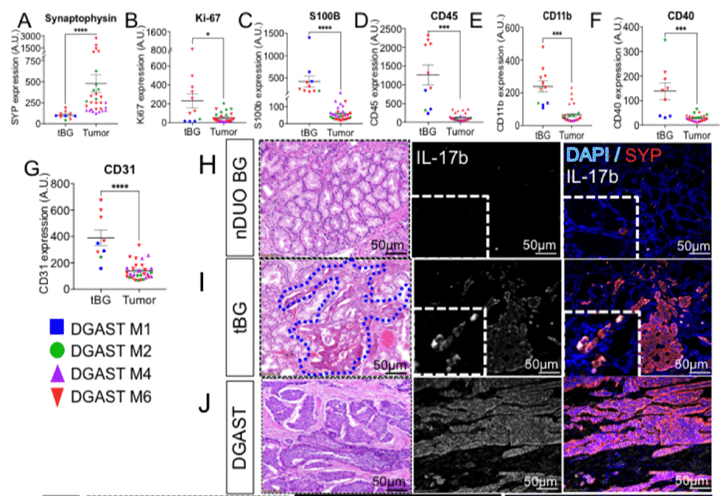
Tumorigenesis and Therapeutic Targets
Combining multiomics and spatial findings, the study determined that NKX6.3 likely plays an active role in tumorigenesis, perhaps through chronic inflammation of normal BG cells. In addition, the identification of inflammatory cytokines in DGASTs suggested cytokine suppression or immune checkpoint inhibitors may be a potential therapeutic path to explore.
As research continues to unravel the mysteries underlying disease development and progression, multiomics and spatial multiomics technologies will continue to provide powerful, informative results driving towards better therapeutic solutions.
BioChain offers a wide selection of FFPE TMAs and FFPE Tissues to be used for control samples in multiomics profiling studies. In addition, if you can’t find the right control samples in our extensive catalogue, BioChain also offers a custom sample collection service to help you obtain samples to meet your specific requirements.
References
- Rico K, Duan S, Pandey RL, et al. Genome analysis identifies differences in the transcriptional targets of duodenal versus pancreatic neuroendocrine tumours. BMJ Open Gastroenterol. 2021;8(1):e000765. doi:10.1136/bmjgast-2021-00076
- Anlauf M, Perren A, Meyer CL, et al. Precursor lesions in patients with multiple endocrine neoplasia type 1-associated duodenal gastrinomas. Gastroenterology. 2005;128(5):1187-1198. doi:10.1053/j.gastro.2005.01.058
Author
Kristi Stephenson received her B.S in Chemistry from the University of Portland. She has over 10 years of experience in the biotech industry, with her primary expertise in sequencing and synthetic biology.

Like this blog? Talk to an expert!
A Peek at a Few of Our Instruments